Cut the phylogeny and continuous trait mapping for a given focal time in the past
Source:R/cut_contMap_for_focal_time.R
cut_contMap_for_focal_time.RdCuts off all the branches of the phylogeny which are
younger than a specific time in the past (i.e. the focal_time).
Branches overlapping the focal_time are shorten to the focal_time.
Likewise, remove continuous trait mapping for the cut off branches
by updating the $tree$maps and $tree$mapped.edge elements.
Arguments
- contMap
Object of class
"contMap", typically generated withphytools::contMap(), that contains a phylogenetic tree and associated continuous trait mapping. The phylogenetic tree must be rooted and fully resolved/dichotomous, but it does not need to be ultrametric (it can includes fossils).- focal_time
Numerical. The time, in terms of time distance from the present, for which the tree and mapping must be cut. It must be smaller than the root age of the phylogeny.
- keep_tip_labels
Logical. Specify whether terminal branches with a single descendant tip must retained their initial
tip.label. Default isTRUE.
Value
The function returns the cut contMap as an object of class "contMap".
It contains a $tree element of classes "simmap" and "phylo". This function updates and adds multiple useful sub-elements to the $tree element.
$mapsAn updated list of named numerical vectors. Provides the mapping of trait values along each remaining edge.$mapped.edgeAn updated matrix. Provides the evolutionary time spent across trait values (columns) along the remaining edges (rows).$root_ageInteger. Stores the age of the root of the tree.$nodes_ID_dfData.frame with two columns. Provides the conversion from thenew_node_IDto theinitial_node_ID. Each row is a node.$initial_nodes_IDVector of character strings. Provides the initial ID of internal nodes. Used to plot internal node IDs as labels withape::nodelabels().$edges_ID_dfData.frame with two columns. Provides the conversion from thenew_edge_IDto theinitial_edge_ID. Each row is an edge/branch.$initial_edges_IDVector of character strings. Provides the initial ID of edges/branches. Used to plot edge/branch IDs as labels withape::edgelabels().
Details
The phylogenetic tree is cut for a specific time in the past (i.e. the focal_time).
When a branch with a single descendant tip is cut and keep_tip_labels = TRUE,
the leaf left is labeled with the tip.label of the unique descendant tip.
When a branch with a single descendant tip is cut and keep_tip_labels = FALSE,
the leaf left is labeled with the node ID of the unique descendant tip.
In all cases, when a branch with multiple descendant tips (i.e., a clade) is cut, the leaf left is labeled with the node ID of the MRCA of the cut-off clade.
The continuous trait mapping is updated accordingly by removing mapping associated with the cut off branches.
Examples
# ----- Prepare data ----- #
# Load mammals phylogeny and data from the R package motmot (data included in deepSTRAPP)
# Initial data source: Slater, 2013; DOI: 10.1111/2041-210X.12084
data(mammals, package = "deepSTRAPP")
mammals_tree <- mammals$mammal.phy
mammals_data <- setNames(object = mammals$mammal.mass$mean,
nm = row.names(mammals$mammal.mass))[mammals_tree$tip.label]
# Run a stochastic mapping based on a Brownian Motion model
# for Ancestral Trait Estimates to obtain a "contMap" object
mammals_contMap <- phytools::contMap(mammals_tree, x = mammals_data,
res = 100, # Number of time steps
plot = FALSE)
# Set focal time
focal_time <- 80
# ----- Example 1: keep_tip_labels = TRUE ----- #
# Cut contMap to 80 Mya while keeping tip.label
# on terminal branches with a unique descending tip.
updated_contMap <- cut_contMap_for_focal_time(contMap = mammals_contMap,
focal_time = focal_time,
keep_tip_labels = TRUE)
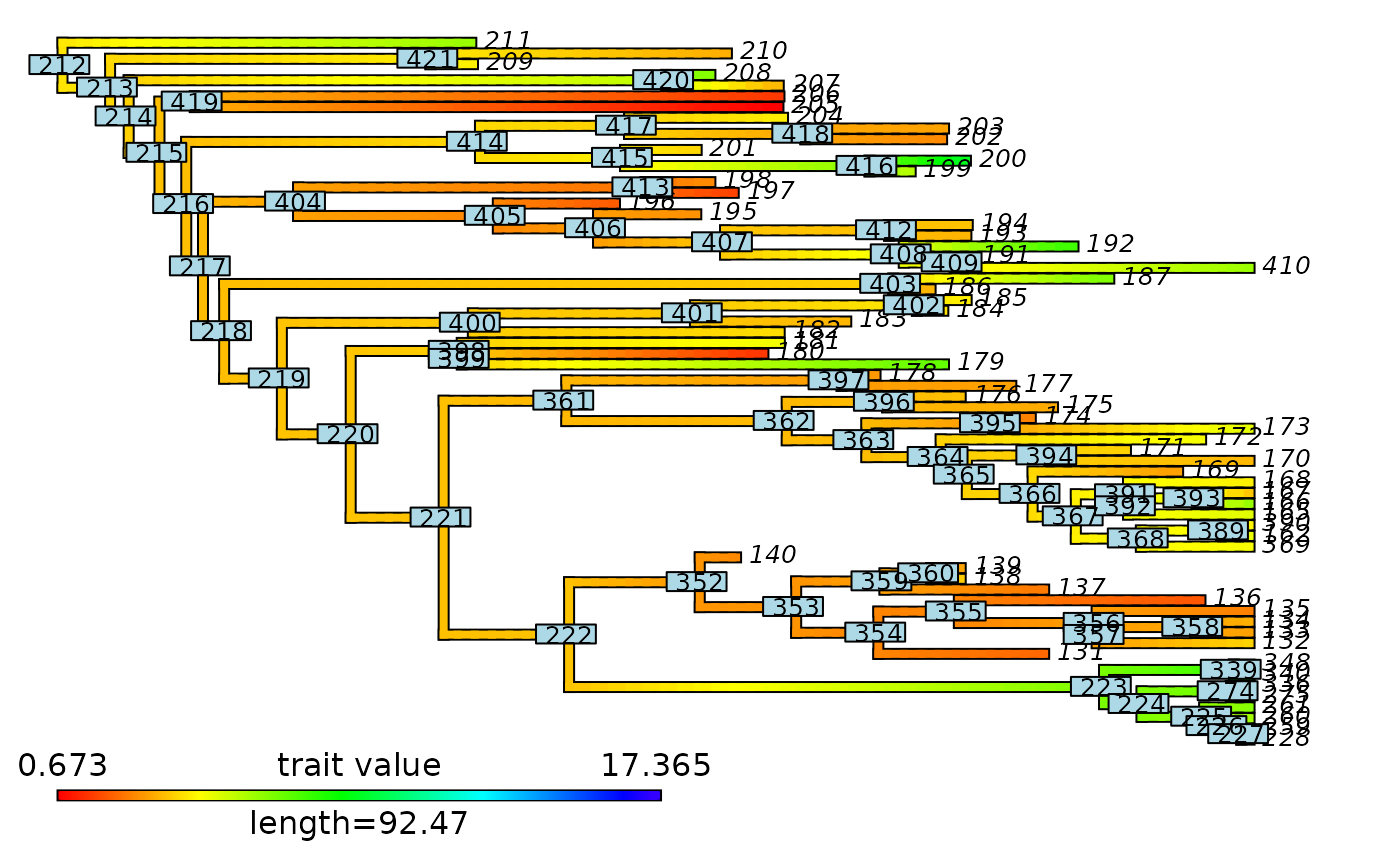
# Plot node labels on initial stochastic map with cut-off
plot_contMap(mammals_contMap, lwd = 2, fsize = c(0.5, 1))
ape::nodelabels(cex = 0.5)
abline(v = max(phytools::nodeHeights(mammals_contMap$tree)[,2]) - focal_time,
col = "red", lty = 2, lwd = 2)
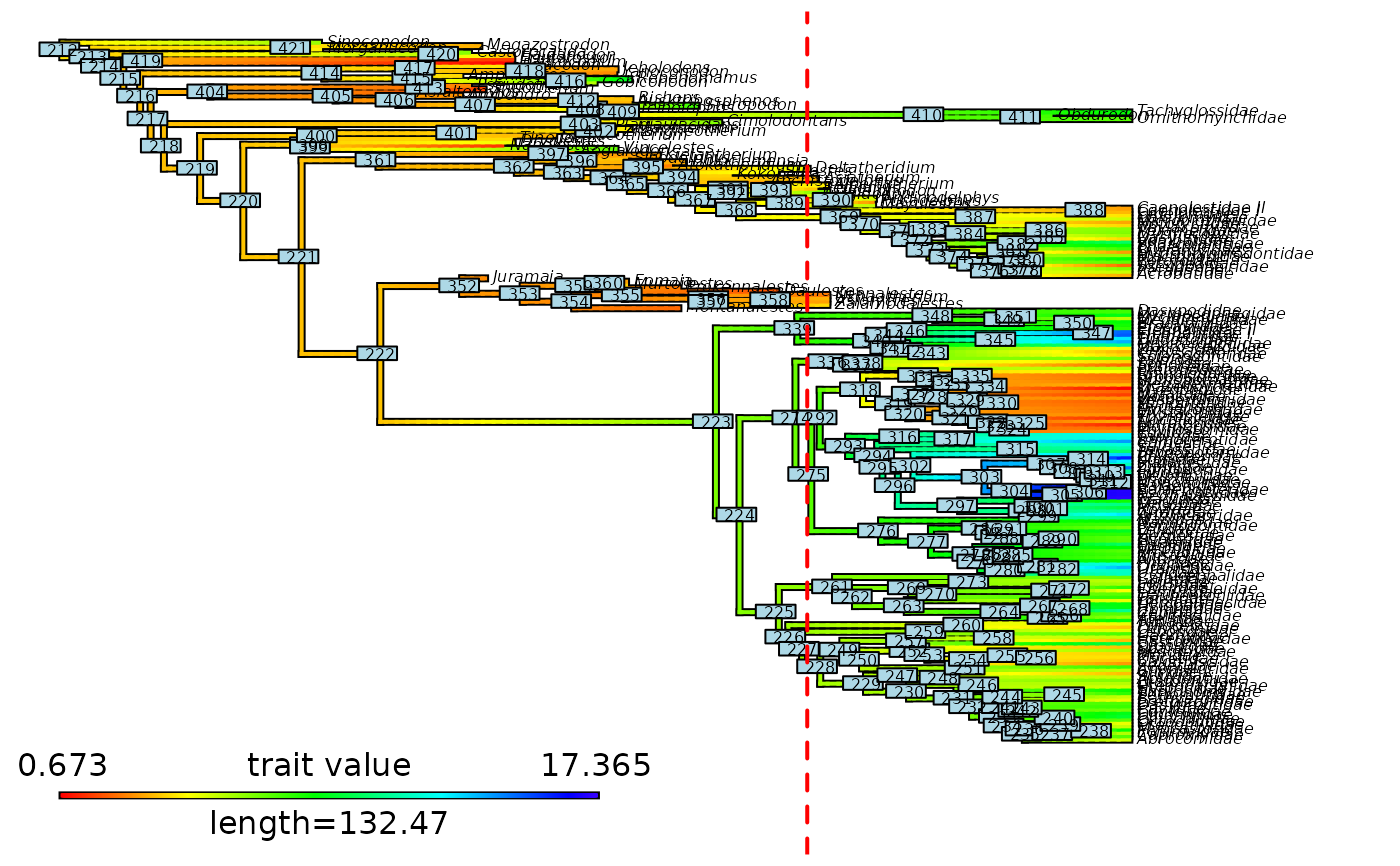 # Plot initial node labels on cut stochastic map
plot_contMap(updated_contMap, fsize = c(0.8, 1))
ape::nodelabels(cex = 0.8, text = updated_contMap$tree$initial_nodes_ID)
# Plot initial node labels on cut stochastic map
plot_contMap(updated_contMap, fsize = c(0.8, 1))
ape::nodelabels(cex = 0.8, text = updated_contMap$tree$initial_nodes_ID)
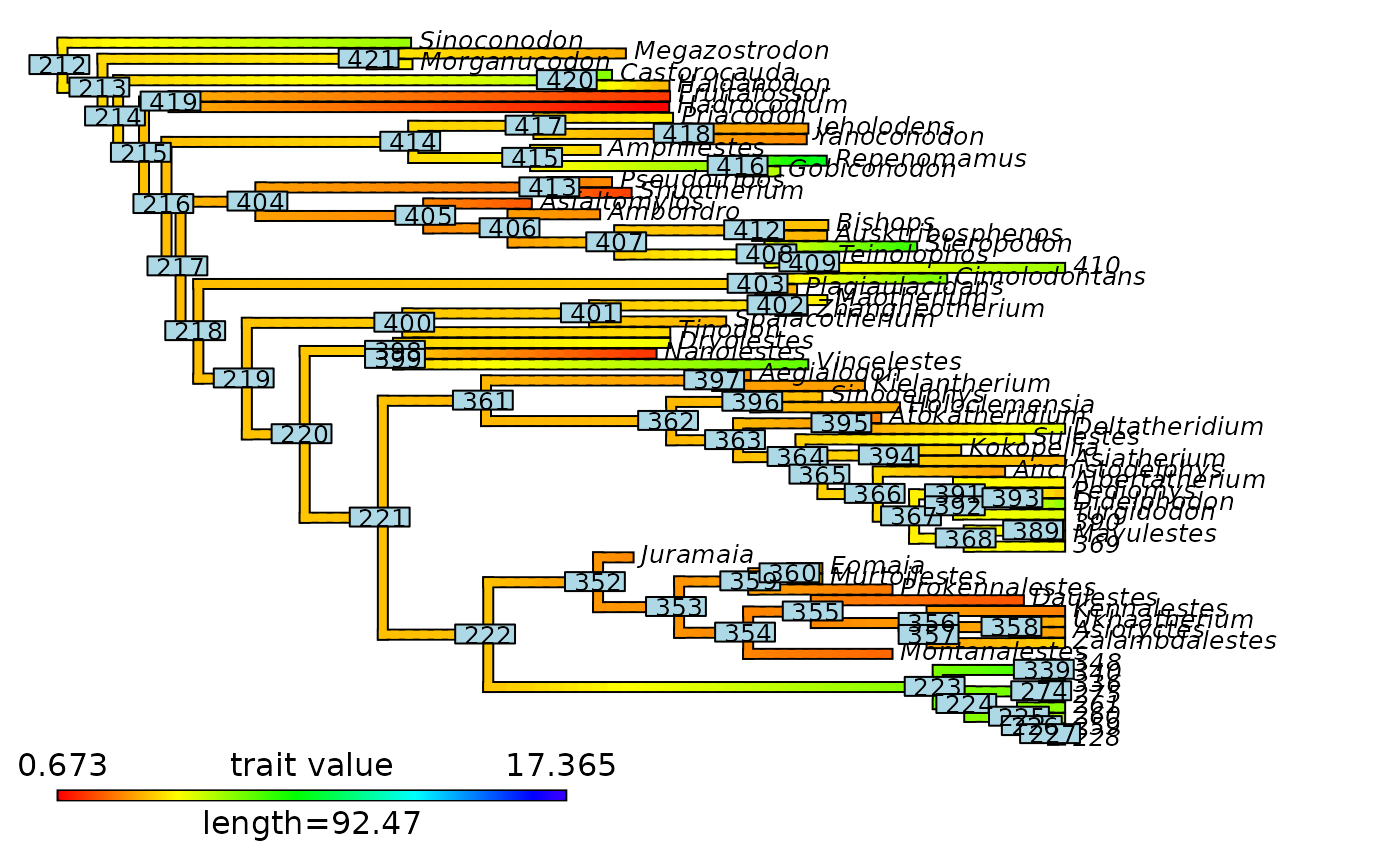 # ----- Example 2: keep_tip_labels = FALSE ----- #
# Cut contMap to 80 Mya while NOT keeping tip.label.
updated_contMap <- cut_contMap_for_focal_time(contMap = mammals_contMap,
focal_time = focal_time,
keep_tip_labels = FALSE)
# Plot node labels on initial stochastic map with cut-off
plot_contMap(mammals_contMap, fsize = c(0.5, 1))
ape::nodelabels(cex = 0.5)
abline(v = max(phytools::nodeHeights(mammals_contMap$tree)[,2]) - focal_time,
col = "red", lty = 2, lwd = 2)
# ----- Example 2: keep_tip_labels = FALSE ----- #
# Cut contMap to 80 Mya while NOT keeping tip.label.
updated_contMap <- cut_contMap_for_focal_time(contMap = mammals_contMap,
focal_time = focal_time,
keep_tip_labels = FALSE)
# Plot node labels on initial stochastic map with cut-off
plot_contMap(mammals_contMap, fsize = c(0.5, 1))
ape::nodelabels(cex = 0.5)
abline(v = max(phytools::nodeHeights(mammals_contMap$tree)[,2]) - focal_time,
col = "red", lty = 2, lwd = 2)
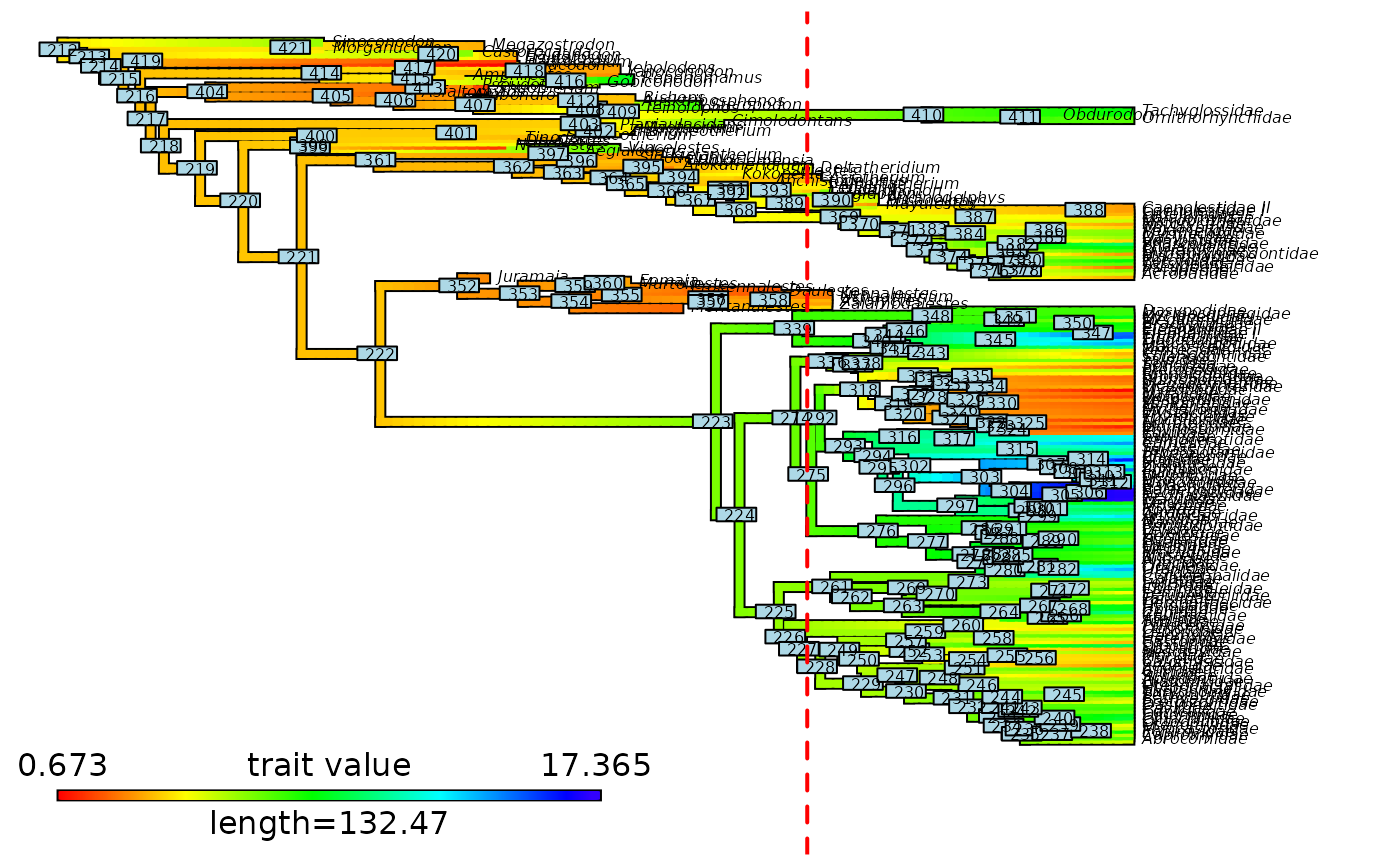 # Plot initial node labels on cut stochastic map
plot_contMap(updated_contMap, fsize = c(0.8, 1))
ape::nodelabels(cex = 0.8, text = updated_contMap$tree$initial_nodes_ID)
# Plot initial node labels on cut stochastic map
plot_contMap(updated_contMap, fsize = c(0.8, 1))
ape::nodelabels(cex = 0.8, text = updated_contMap$tree$initial_nodes_ID)